A design‐by‐treatment interaction model for network meta‐analysis and meta‐regression with integrated nested Laplace approximations
Abstract
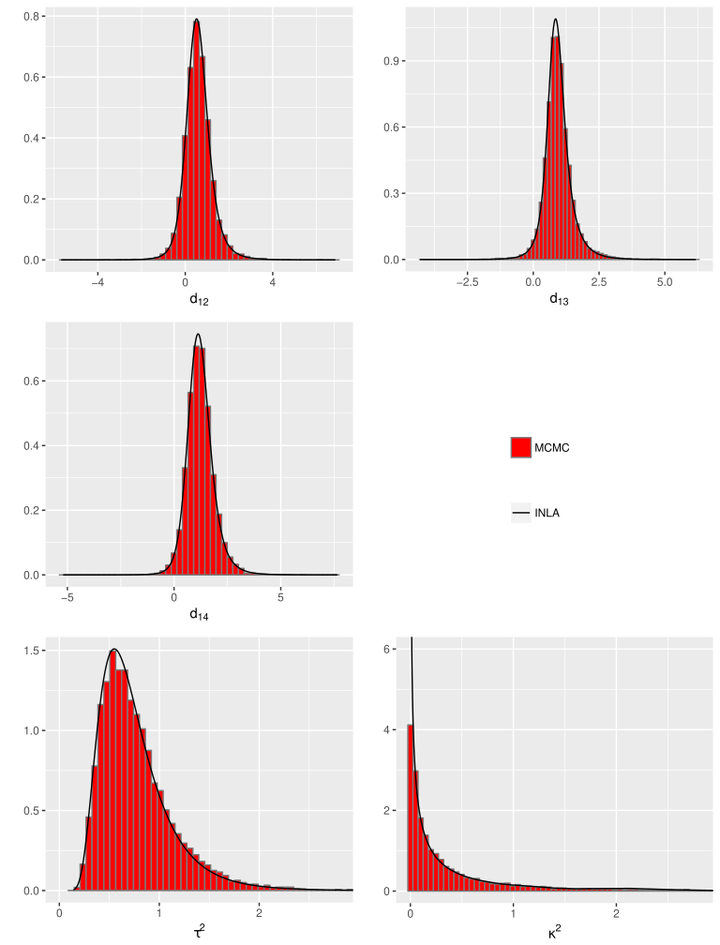
Network meta‐analysis (NMA) is gaining popularity for comparing multiple treatments in a single analysis. Generalized linear mixed models provide a unifying framework for NMA, allow us to analyze datasets with dichotomous, continuous or count endpoints, and take into account multiarm trials, potential heterogeneity between trials and network inconsistency. To perform inference within such NMA models, the use of Bayesian methods is often advocated. The standard inference tool is Markov chain Monte Carlo (MCMC), which is computationally expensive and requires convergence diagnostics. A deterministic approach to do fully Bayesian inference for latent Gaussian models can be achieved by integrated nested Laplace approximations (INLA), which is a fast and accurate alternative to MCMC. We show how NMA models fit in the class of latent Gaussian models and how NMA models are implemented using INLA and demonstrate that the estimates obtained by INLA are in close agreement with the ones obtained by MCMC. Specifically, we emphasize the design‐by‐treatment interaction model with random inconsistency parameters (also known as the Jackson model). Also, we have proposed a network meta‐regression model, which is constructed by incorporating trial‐level covariates to the Jackson model to explain possible sources of heterogeneity and/or inconsistency in the network. A publicly available R package, nmaINLA, is developed to automate the INLA implementation of NMA models, which are considered in this paper. Three applications illustrate the use of INLA for a NMA.